Vibrational-Rotational Spectroscopy of HCl
Author: James M. McCormick
Last Update: February 5, 2014
Introduction
In this exercise you will be evaluating whether the Morse potential is a sufficiently good model for the single vibration of the HCl molecule. You will be acquiring IR absorbance data from which you will extract the relevant parameters and you will also be using computational chemistry methods to exam an alternate way of modeling the H-Cl bond. To obtain the most precise values possible for these parameters, you will need to acquire data for two different isotopomers of HCl. While our FTIR spectrometers can resolve the peaks arising from the 1H-35Cl and 1H-37Cl isotopomers, the instrument’s precision is not high enough for our analysis. Therefore, you will also be acquiring data for the 2H-35Cl and 2H-37Cl isotopomers. The handouts,1,2 your textbook,3 and the online reference4 give more background on the theory and the methods that you will use to extract the rotational and vibrational parameters from your data. You will follow the data analysis method given in reference 1, but a different method will be used to generate a mixture of HCl and DCl,5 so you do not need to be concerned with the procedures given in references 1 and 2. You also do not need to be concerned with any statistical mechanical calculations. The computational component is drawn from a literature procedure.6 This will be modified to some extent, as outlined below, because you will be using the WebMO interface7 for Gaussian.8 For additional information on computational chemistry in general,9 and the Gaussian computational package10 you are referred to the given references. If you would like to review the basic WebMO procedures, please click here to view the CHEM 131 Computational Chemistry Exercise.
Experimental
Laboratory Component
You will be using a gas IR cell fitted with KBr windows in this experiment. Do not remove, touch, or attempt to clean the windows. Gas cells of this type are fragile and very expensive. Do not remove the Tygon tubing from the stopcocks on the cell itself. Disconnect all tubing as directed below. CAUTION! HCl, DCl and SO2 are corrosive, poisonous gases; avoid inhalation. Be familiar with the safe handling procedures for this material before starting work in the lab.
A schematic representation of the apparatus used to generate the mixture of HCl and DCl (and SO2) is shown in Fig. 1. It is modified slightly from a similar apparatus described in the literature.5 The whole apparatus will be flushed with nitrogen before and after use on a Schlenk line in the hood. Click here to view instructions on Schlenk line operations.
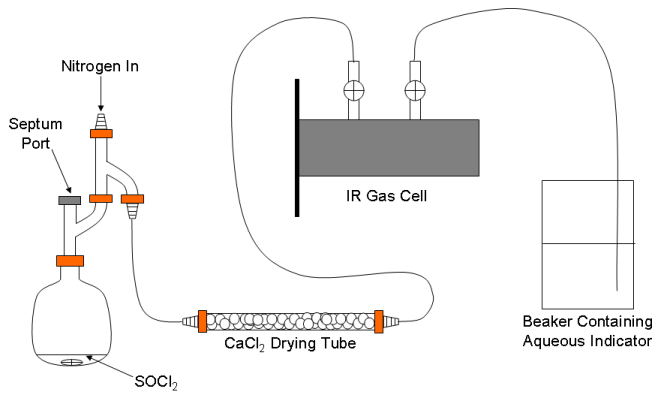
Figure 1. Schematic of the apparatus for generating the gaseous HCl/DCl mixture.
Place 1 mL of SOCl2 and a small magnetic stirring bar in the round bottom flask.5 Assemble the apparatus, but do not connect the IR gas cell yet (leave it in the desiccator for now). Place clamps on the Claisen adapter just above the flask and on the Claisen just below where the nitrogen will enter. Connect the hose barb on the top of the apparatus (labeled “Nitrogen In” in Fig. 1) to one of ports on the Schlenk line with the red rubber vacuum hose already attached to the port. Remove the serum stopper from one end of the drying tube and connect it to the apparatus using a short piece of tubing. Open the septum port and then open the stopcock on the Schlenk line to admit nitrogen to the apparatus. Increase the flow to about 10 bubbles of nitrogen per second passing through the Schlenk line’s input bubbler (there should be little if any nitrogen flow through the Schlenk line’s exhaust bubbler) and start slowly stirring the SOCl2. Purge the system in this way for approximately 10 min. Remove the septum from the other end of the drying tube and replace the septum port. Purge for another 5 min.
Remove the gas cell from the desiccator (use two hands to carefully remove the desiccator’s lid and use two hands to carry the cell). Be mindful of the hoses at all times. Close both of the cell’s stopcocks. Carefully remove the hose nipple from the open end of the drying tube and carefully attach the gas cell’s hose nipple. IMPORTANT! When you do this you will need to reduce the nitrogen flow to about 1 bubble per second at the inlet bubbler. As you attach the cell to the rest of the apparatus you will see a lot of gas being discharged through the Schlenk line’s exhaust bubbler. This is not a problem, unless the flow is so high that mineral oil is splashed out of the bubbler.
Place the end of the other tube attached to the gas cell in a small beaker of water containing a sufficient amount of the indicator so that you can see some color. Once the flow has been returned to a low level open the gas cell’s inlet stopcock and then open the stopcock leading to the indicator solution. You should see bubbles of gas exiting through the solution. If you do not, pull up the end of the tube slightly or increase the flow of nitrogen slightly. If you see water being drawn back into the tube, immediately close the stopcock on the gas cell leading to the solution and seek assistance.
Let nitrogen flow through the gas cell for about 5 min. While this is happening place the hose nipple that you just removed from the drying tube on one of the other Schlenk ports. Check if there is any leftover H2O/D2O mixture from a previous experiment. If there is none, prepare it as described in reference 6.
Take up about 0.5 mL of the H2O/D2O mixture into a 1 mL syringe equipped with a pipetting needle. Insert the needle through the septum port and slowly add the H2O/D2O mixture to the SOCl2. Do not get the needle too close to the SOCl2 and do not add the H2O/D2O mixture so quickly that the reaction splashes out of the flask. You will notice that there will be an immediate and vigorous evolution of gas. When the indicator solution changes color, remove the needle. Immediately close the stopcock on the gas cell leading to the indicator solution and quickly remove the end of this tube from the solution. If any water has been sucked back into the tube carefully remove it and dry the tube. Do not get water anywhere near the IR cell windows! Close the gas cell’s other stopcock and disconnect the cell from the drying tube. The cell is now filled with a mixture of HCl, DCl and SO2 and is ready for the measurement. You can leave nitrogen flowing through the apparatus and out through the drying tube as you begin data acquisition. However, you must thoroughly rinse the syringe and the needle with distilled water and then with methanol within a few minutes of finishing the addition of the H2O/D2O mixture to the SOCl2.
You will be collecting data using the Nicolet Avatar FTIR in the Advanced Laboratory Instrument room running the OMNIC software package. Be sure to start OMNIC, not EZ OMNIC (both icons are on the desktop, but EZ OMNIC has limited capabilities). To set up the spectrometer to acquire data select Collect, Experiment Setup from the menu bar. The experiment setup window (similar to the one shown in Fig. 2) will appear. Set the parameters as shown in Fig. 2 and click OK.
Figure 2. Experiment setup window in the OMNIC software package.
To start data acquisition click on the icon labeled Col Smp just below the menu bar. You will be prompted to obtain a background. You can just click OK to obtain the background with an empty sample chamber. Once the background has been acquired, the software will prompt you that it is ready to obtain the spectrum. Place the gas cell filled with the HCl/DCl mixture in the cell holder (note that the rectangular metal piece slides into the instrument’s sample holder) and click OK. Once the instrument is finished acquiring the spectrum you will be prompted to enter a title (enter something appropriate and click OK). You will then be asked whether you want to add the spectrum to window 1. In most cases, the answer isYes. Save the spectrum in the standard Nicolet format and as an ASCII text file (CSV format) for importing into Excel later. Use OMNIC’s peak picking routine to determine each transition’s wavenumber. This can also be done in LoggerPro (simple cut and paste the data from the Excel file into the data window in LoggerPro). It is advised that you not try to do the peak picking in Excel as it is extremely tedious.
Once data acquisition has finished, bring the cell back to the hood. Reconnect the drying tube to the cell and then disconnect the drying tube from the apparatus. Connect the drying tube to the other Schlenk port (vide supra). Close the stopcock on the manifold leading to the apparatus and open the stopcock leading to the drying tube. Open the stopcock on the gas cell leading to the drying tube. Open the other stopcock (the tube does not need to be in water at this point, but should be directed toward the back of the hood). Flush the cell for at least 10 min and then test whether the gas contains any acid by placing the end of the exhaust tube in a fresh indicator solution. Continue purging until the indicator no longer changes color.
While the cell is being purged, carefully disassemble the apparatus. Place the flask containing the SOCl2 in a beaker in the back of the hood. Carefully add small portions of water to destroy any remaining SOCl2. Discard the neutralized solution in a drain with copious amounts of water. Thoroughly rinse the apparatus with distilled water, then with methanol and allow to air dry.
Once the cell is completely purged of the acidic gases, remove the open tube from the indicator solution and close the stopcock on the gas cell leading to it. Close the gas cell’s other stopcock and disconnect the drying tube. Reconnect the original hose nipple to the drying tube. Shut off the nitrogen flow and remove the drying tube from the Schlenk line. Place the serum stoppers over the hose nipples and return the drying tube to the desiccator. Open both stopcocks on the gas cell and carefully return the gas cell to the desiccator.
Computational Component
Click here to proceed to the Truman WebMO7 site which is our portal for Gaussian.8 The easiest way to generate the input file is to first create a new job and use the molecule editor to draw it. To review how to do this, click here to open the CHEM 131 laboratory exercise, or click here to view the PDF version of the WebMO User’s Guide (click on the Bookmarks tab for easier navigation in this file). Once you have drawn the molecule, you will proceed to the Configure Job Options Screen. Enter a name for your job and then click on the Preview tab. Now click on the Generate button. If you drew HCl, then the text displayed in Fig. 3 will be displayed. This is an editable view of the actual input file that Gaussian will read as it starts your calculation. The different sections of the input file are explained in reference 6. To perform the calculations for He and HCl described in reference 6, you will need to change the Route Section (everything after the #N on the first line). When you make these changes be sure to pay close attention to spaces and blank lines; they are very important and a misplaced space can lead to very unexpected results. Perform the calculations described for He in reference 6 before attempting the HCl calculations as this will allow you to check whether you are performing the calculations correctly. Note that you do not need to discuss the He results in the main body of the report, but the results and some analysis should be included in the supplemental material. If you wish, you can set up the HCl coordinate scan to generate more points than the nine indicated in reference 6. However, if you do so you should retain the limits of the scan.
#N HF/6-311+G(d,p) OPT FREQ
HCl Hartree-Fock calculation
0 1
Cl
H 1 B1
B1 1.2694990
Figure 3. A typical Gaussian input file for HCl that will perform a geometry optimization followed by a vibrational frequency calculation using Hartree-Fock theory and the 6-311+G(d,p) basis set.
Once a calculation is complete, the output file (called the archive in reference 6) may be viewed by clicking on the Raw Output link on the View Job page for the calculation. This is simply a text file from which you can copy and paste as you normally do.
Analysis and Discussion
Perform the analysis described in reference 1 for HCl and DCl (if you want you can also perform it for the isotopomers containing different Cl isotopes). Be sure that you have correctly assigned which peaks come from which isotopomer and correctly indexed the peaks as described.1 Prepare a graph of wavenumber as a function of m for both the 1H-35Cl and the 2H-35Cl peaks as this will help you find any errors. Only perform the cubic fit on both data sets. Propagate the error in the transition energy through to the final results.
Your results section should have a figure showing the entire spectrum (in absorbance units) and indicating which peaks arise from which species, as well as separate figures showing only the 1H-35Cl transitions and only the 2H-35Cl transitions. Remember that, even though a spectrum is experimentally determined, it is traditional to show spectra as solid lines. There must also be a figure showing your fit of the wavenumber of each 1H-35Cl transition as a function of m.1 There must also be a representative figure showing the fit of the computational results as described in reference 6.
Tabulate the experimental (and the uncertainty at 95% confidence) and theoretical values for each spectroscopic parameter. Compare the experimental results with the theoretical results and your results with those in the literature. Discuss any relevant differences.
1. Shoemaker, D. P.; Garland, C. W. and Nibler, J. W. Experiments in Physical Chemistry, 7thEd.; McGraw-Hill: New York, 2003; p. 403-411.
2. Halpern, A. M. and McBane, G. C. Experimental Physical Chemistry: A Laboratory Textbook, 3rd Ed.; Freeman: New York, 2006; p. 36.1-36.14.
3. Atkins, P. W. and de Paula, J. Physical Chemistry, 8th Ed., W. H. Freeman and Co.: New York, 2006, p. 441-462.
4. Stafford, F. E.; Holt, C. W. and Paulson, G. L. J. Chem. Educ. 1963, 40, 245. Click here to view this article (Truman addresses and J. Chem. Educ. subscribers only).
5. Mayer, S. G.; Bard, R. B. and Cantrell, K. J. Chem. Educ. 2008, 85, p. 847-848. Click hereto view this article (Truman addresses and J. Chem. Educ. subscribers only).
6. Halpern, A. M. and McBane, G. C. Experimental Physical Chemistry: A Laboratory Textbook, 3rd Ed.; Freeman: New York, 2006; p. 41.1-41.8.
7a. Polik, W. F and Schmidt, J. R. WebMO Pro 6.1.010p; WebMO LLC: Holland, MI, 2006.
b. Polik, W. F. and Schmidt, J. R. WebMO User’s Guide; 2003,http://www.webmo.net/download/WebMO_Users_Guide.pdf.
8. Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Montgomery, Jr., J. A.; Vreven, T.; Kudin, K. N.; Burant, J. C.; Millam, J. M.; Iyengar, S. S.; Tomasi, J.; Barone, V.; Mennucci, B.; Cossi, M.; Scalmani, G.; Rega, N.; Petersson, G. A.; Nakatsuji, H.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Klene, M.; Li, X.; Knox, J. E.; Hratchian, H. P.; Cross, J. B.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R. E.; Yazyev, O.; Austin, A. J.; Cammi, R.; Pomelli, C.; Ochterski, J. W.; Ayala, P. Y.; Morokuma, K.; Voth, G. A.; Salvador, P.; Dannenberg, J. J.; Zakrzewski, V. G.; Dapprich, S.; Daniels, A. D.; Strain, M. C.; Farkas, O.; Malick, D. K.; Rabuck, A. D.; Raghavachari, K.; Foresman, J. B.; Ortiz, J. V.; Cui, Q.; Baboul, A. G.; Clifford, S.; Cioslowski, J.; Stefanov, B. B.; Liu, G.; Liashenko, A.; Piskorz, P.; Komaromi, I.; Martin, R. L.; Fox, D. J.; Keith, T.; Al-Laham, M. A.; Peng, C. Y.; Nanayakkara, A.; Challacombe, M.; Gill, P. M. W.; Johnson, B.; Chen, W.; Wong, M. W.; Gonzalez, C. and Pople, J. A.Gaussian 03, Revision C.02; Gaussian, Inc.: Wallingford, CT, 2004.
9. Hehre, W. J. in Engel, T. and Reid, P. Physical Chemistry; Benjamin Cummings: San Francisco, CA, 2006; p. 597-655.
10. Forseman, J. B. and Frisch, A. Exploring Chemistry with Electronic Structure Methods: A Guide to Using Gaussian; Gaussian, Inc.: Pittsburgh, PA, 1993.
